EpiCypher是一家為表觀遺傳學(xué)和染色質(zhì)生物學(xué)研究提供高質(zhì)量試劑和工具的專業(yè)制造商。EpiCypher推出的CUT&RUN級別的Histone H3K4me2 Antibody符合EpiCypher的批次特異性SNAP-CertifiedTM標(biāo)準(zhǔn)����,在CUT&RUN中具有特異性和高效的靶點富集。通過SNAP-CUTANA?K-MetStat Panel的spike-in對照確定(EpiCypher 19-1002����,F(xiàn)ig. 1)與相關(guān)組蛋白PTM的交叉反應(yīng)性<20%,在500k和50k起始細胞中一致的基因組富集證實了高靶向效率(Fig. 2-4)�����。該抗體靶向的H3K4me2在轉(zhuǎn)錄活性基因的啟動子和細胞發(fā)育過程中表達的基因中富集[1]。
[1] Pekowkska et al. Genome Res. (2010). PMID: 20841431.
產(chǎn)品詳情
反應(yīng)種屬: Human, Mouse, Wide Range (Predicted)
宿主來源: Rabbit
實驗應(yīng)用: CUT&RUN, ICC, WB
免疫原: A synthetic peptide corresponding to histone H3 dimethylated at lysine 4
克隆性: Monoclonal
表達系統(tǒng): HEK293 cells
保存溫度: Stable for 1 year at -20°C from date of receipt
運輸溫度: Frozen cold packs.
產(chǎn)品形式: Protein A affinity-purified antibody in PBS, 0.09% sodium azide, 1% BSA, and 50% glycerol
驗證數(shù)據(jù)
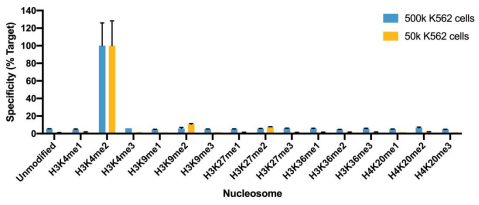
Figure 1: SNAP specificity analysis in CUT&RUN
CUT&RUN was performed as described above. CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-MetStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K4me2 set to 100%).
?
?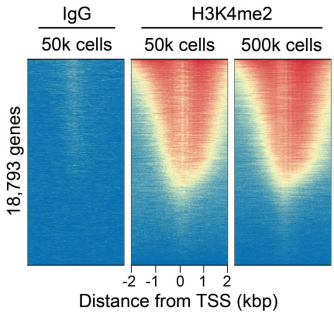
Figure 2: CUT&RUN genome wide enrichment
CUT&RUN was performed as described above. Sequence reads were aligned to 18,793 annotated transcription start sites (TSSs ± 2 kbp). Signal enrichment was sorted from highest to lowest (top to bottom) relative to the H3K4me2 - 50k cells sample (all gene rows aligned). High, medium, and low intensity are shown in red, yellow, and blue, respectively. H3K4me2 antibody produced the expected TSS enrichment pattern, which was consistent between 500k and 50k cells and greater than the IgG negative control.
 ?
?
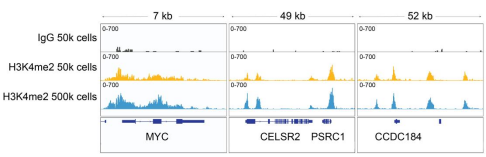
Figure 3: H3K4me2 CUT&RUN representative browser tracks
CUT&RUN was performed as described above. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). Similar results in peak structure and location were observed for both cell inputs.
 ?
?
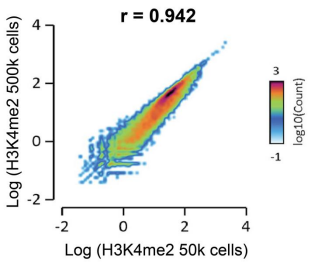
Figure 4: Antibody efficiency analysis in CUT&RUN using cell input correlation
CUT&RUN was performed as described above. Genome-wide correlation analysis was performed to compare H3K4me2 antibody enrichment using 500k cell and 50k cell inputs. The log of the number of reads per 75 bp binned region across the genome is plotted for both samples. CUT&RUN data generated using this H3K4me2 antibody are highly correlated between the two cell inputs (Pearson correlation r = 0.942), indicating high efficiency of H3K4me2 antibody target recovery.
 ?
?
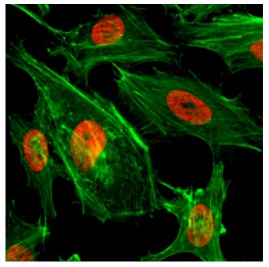
Figure 5: Immunocytochemistry
ICC of HeLa cells using 2 μg/mL of H3K4me2 antibody (red). Actin filaments were labeled with fluorescein phalloidin (green).
 ?
?
Figure 6: Western blot data
Recombinant histone H3.3 (Lane 1) and acid extracts of HeLa cells (Lane 2) were blotted onto PVDF and probed with 0.025 μg/mL of H3K4me2 antibody.
訂購詳情
貨號 | 產(chǎn)品名稱 | 規(guī)格 |
13-0027 | Histone H3K4me2 Antibody, SNAP-Certified? for CUT&RUN | 100 μg |

如需了解更多詳細信息或相關(guān)產(chǎn)品����,請聯(lián)系EpiCypher中國授權(quán)代理商-欣博盛生物?
全國服務(wù)熱線: 4006-800-892 ? ? ??郵箱: market@neobioscience.com?
深圳: 0755-26755892 ? ? ? ??北京: 010-88594029 ???????????
廣州:020-87615159????????? ?上海: 021-34613729
代理品牌網(wǎng)站: m.smblzp.com?
自主品牌網(wǎng)站: www.neobioscience.net