越來(lái)越多的證據(jù)表明�����,核小體是體外表征許多染色質(zhì)調(diào)節(jié)因子的最佳底物��。隨著攜帶完全確定組蛋白修飾的重組核小體的出現(xiàn)��,為下一代染色質(zhì)研究提供了新的或更好的方法(如染色質(zhì)結(jié)合蛋白分析���、酶分析、抗體譜分析)�����。EpiCypher擁有令人印象深刻的產(chǎn)品目錄�,其中包括83個(gè)獨(dú)特的重組核小體庫(kù)存,并且有能力生產(chǎn)定制設(shè)計(jì)核小體��,質(zhì)量高、周期短�,助力您的研究!
?
在科研實(shí)驗(yàn)中���,如果您要使用重組核小體進(jìn)行染色質(zhì)實(shí)驗(yàn)��,則需考慮以下幾點(diǎn):
?
1�、用于開(kāi)發(fā)修飾組蛋白的方法�����。確保使用無(wú)疤痕方法整合所需的PTMs是很重要的�,這種方法可以重現(xiàn)天然組蛋白結(jié)構(gòu)。EpiCypher 使用幾種不同的方法獲得修飾的組蛋白�,所有方法都會(huì)無(wú)疤痕整合組蛋白修飾。
為什么這很重要呢�����?許多市售的重組核小體是使用組蛋白PTM類(lèi)似物構(gòu)建的�,如甲基賴(lài)氨酸類(lèi)似物(MLAs) 10,其會(huì)導(dǎo)致修飾位點(diǎn)的氨基酸序列發(fā)生變化��。這些非天然組蛋白修飾類(lèi)似物已被證明會(huì)破壞與染色質(zhì)調(diào)節(jié)蛋白和組蛋白PTM特異性抗體的相互作用���,并且這對(duì)研究生理機(jī)制來(lái)講是不理想的���。因此�,使用這些方法合成的核小體時(shí)應(yīng)格外謹(jǐn)慎11-13�����。
?
2�����、修飾組蛋白的純度����。組裝完全定義的同質(zhì)核小體的下一步是對(duì)修飾組蛋白進(jìn)行嚴(yán)格的質(zhì)量控制。
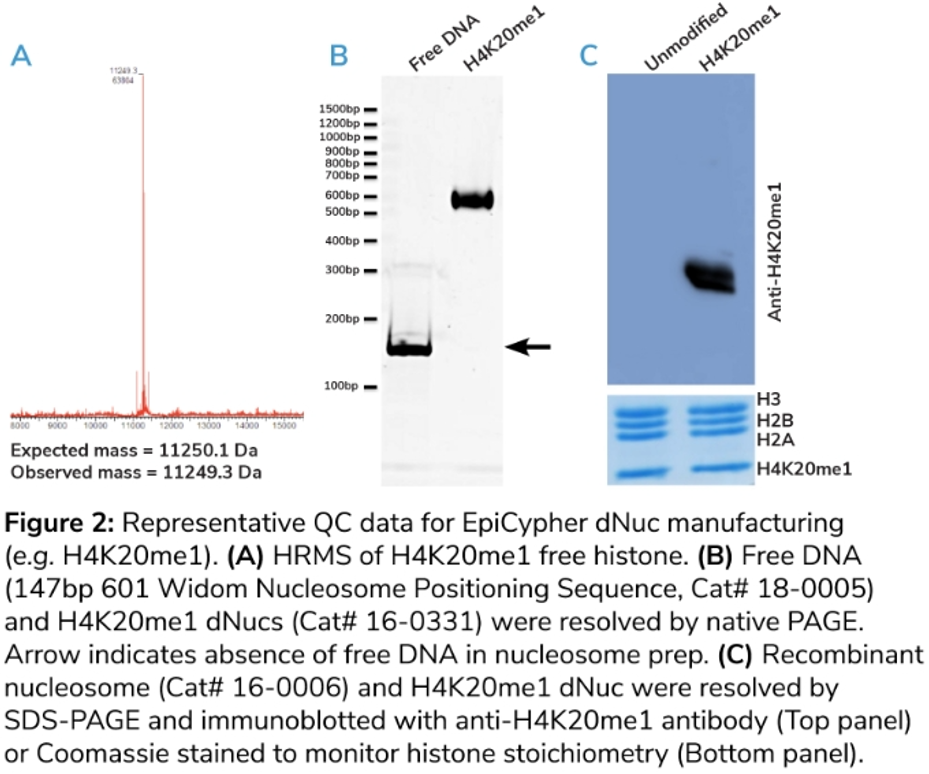
在質(zhì)控結(jié)果中�����,HPLC 跡線應(yīng)顯示是單一洗脫物質(zhì)�,表明組蛋白純度>95%��;平行高分辨率質(zhì)譜(HRMS)應(yīng)在預(yù)期質(zhì)量的1道爾頓范圍內(nèi)顯示一個(gè)單峰�,沒(méi)有任何意義的額外電荷質(zhì)量(m/z)信號(hào)(例如圖2A)����。EpiCypher的所有修飾組蛋白均通過(guò)HPLC和HRMS分析進(jìn)行驗(yàn)證�。
為什么這很重要呢?不必要的物質(zhì)(如甲硫氨酸氧化)可能引起結(jié)構(gòu)改變�����,并影響靜電相互作用或疏水相互作用�����,從而損害下游核小體組裝的效率�。
?
?
3、組裝核小體的質(zhì)量控制(QC)指標(biāo)�。DNA組裝后核小體的質(zhì)量驗(yàn)證對(duì)最終產(chǎn)品的信任保證至關(guān)重要。?
EpiCypher使用天然PAGE分析DNA上的dNuc組裝��,其中使用約150bp DNA的高效組裝應(yīng)該只產(chǎn)生單一物質(zhì)��,這相對(duì)于未組裝的游離DNA���,其遷移率降低(圖2B��,下圖)���。
為什么這很重要呢����?因?yàn)楸晃廴镜挠坞xDNA會(huì)誘導(dǎo)染色質(zhì)修飾酶(如NSD2)的異?���;钚裕员仨毐苊?�。此外���,次優(yōu)組裝可能導(dǎo)致樣品的異質(zhì)性混合�,包括錯(cuò)誤定位的核小體����。
我們還使用考馬斯染色對(duì)最終的dNucs進(jìn)行SDS-PAGE分析,以確保四種組蛋白的化學(xué)計(jì)量相等(圖2C�,下圖)。然后�����,我們通過(guò)免疫印跡證實(shí)了整合組蛋白PTM的存在(圖2C��,上圖)����。
為什么這很重要呢?如果其他種類(lèi)的蛋白質(zhì)污染或偏離1:1:1:1的比例�,則可能表明組蛋白降解或組裝不良,因此有必要重新解析每個(gè)組蛋白�。而免疫印跡對(duì)于確定修飾是否存在于組蛋白的正確位置上非常重要。
?
核小體相關(guān)產(chǎn)品
分類(lèi) | 貨號(hào) | 產(chǎn)品名稱(chēng) |
rNucs Human Recombinant? Nucleosomes, No PTMs | 16-0006 | Mononucleosomes, biotinylated |
16-0009 | Mononucleosomes, non-biotinylated |
16-0024 | Mononucleosomes, desthiobiotinylated |
16-0027 | Tailless Nucleosomes, biotinylated |
16-0023 | Mononucleosomes, H3.1 ΔN2, biotinylated |
16-0016 | Mononucleosomes, H3.1 ΔN32, biotinylated |
16-1016 | Mononucleosomes, H3.1 ΔN32, non-biotinylated |
16-0017 | Mononucleosomes, H3.3 ΔN32, biotinylated |
16-1017 | Mononucleosomes, H3.3 ΔN32, non-biotinylated |
16-0018 | Mononucleosomes, H4 ΔN15, biotinylated |
16-3004 | Dinucleosomes, biotinylated |
16-3104 | Dinucleosomes, non-biotinylated |
|
|
|
dNucs Designer Recombinant? Nucleosomes with PTMs | 16-0321 | H3K4me1, biotinylated |
16-0334 | H3K4me2, biotinylated |
16-1334 | H3K4me2, non-biotinylated |
16-0316 | H3K4me3, biotinylated |
16-1316 | H3K4me3, non-biotinylated |
16-0402 | H3K4,K9me3, biotinylated |
16-0403 | H3K4,K27me3, biotinylated |
16-0335 | H3K4me3,K9,14,18ac, biotinylated |
16-0325 | H3K9me1, biotinylated |
16-0324 | H3K9me2, biotinylated |
16-0315 | H3K9me3, biotinylated |
16-0338 | H3K27me1, biotinylated |
16-0339 | H3K27me2, biotinylated |
16-0317 | H3K27me3, biotinylated |
16-1317 | H3K27me3, non-biotinylated |
16-0397 | H3.1K27me3,S28phos, biotinylated |
16-0322 | H3K36me1, biotinylated |
16-0319 | H3K36me2, biotinylated |
16-0320 | H3K36me3, biotinylated |
16-1320 | H3K36me3, non-biotinylated |
16-0390 | H3.3K36me3, biotinylated |
16-0367 | H3K79me1, biotinylated |
16-0368 | H3K79me2, biotinylated |
16-0369 | H3K79me3, biotinylated |
16-0393 | H4K12me1, biotinylated |
16-0331 | H4K20me1, biotinylated |
16-0332 | H4K20me2, biotinylated |
16-0333 | H4K20me3, biotinylated |
16-1333 | H4K20me3, non-biotinylated |
|
|
|
vNucs Histone Variants | 16-0013 | H2AX, biotinylated |
16-1013 | H2AX, non-biotinylated |
16-0366 | H2AXS139phos, biotinylated |
16-0014 | H2AZ.1, biotinylated |
16-1014 | H2AZ.1, non-biotinylated |
16-0015 | H2AZ.2, biotinylated |
16-0011 | H3.3, biotinylated |
16-0012 | H3.3, non-biotinylated |
|
|
|
Mutant Nucs Defined Amino? Acid Substitutions | 16-0029 | H2AE61A, biotinylated |
16-1029 | H2AE61A, non-biotinylated |
16-0030 | H2AE92K, biotinylated |
16-1030 | H2AE92K, non-biotinylated |
16-0031 | H2BE105A,E113A, biotinylated |
16-1031 | H2BE105A,E113A, non-biotinylated |
16-0349 | Oncogenic Nucs (oncoNucs) |
16-0350 | H3.3K9M, biotinylated |
16-1323 | H3.3K27M, biotinylated |
16-0323 | H3.3K27M, non-biotinylated |
16-0346 | H3.3G34R, biotinylated |
16-0347 | H3.3G34V, biotinylated |
16-0348 | H3.3G34W, biotinylated |
16-0344 | H3.3K36M, biotinylated |
|
|
|
Methyl DNA Nucs Nucleosomes? with Methylated DNA | 16-2043 | Mononucleosomes, Recombinant,? Hemi-methylated 199x601 DNA, biotinylated |
16-2143 | Mononucleosomes, Recombinant,? Hemi-methylated 199x601 DNA, non-biotinylated |
16-2044 | Mononucleosomes, Recombinant,? 199x601 DNA, biotinylated |
16-2144 | Mononucleosomes, Recombinant,? 199x601 DNA, non-biotinylated |
16-2045 | Mononucleosomes, Recombinant, Symmetrically? Methylated 199x601 DNA, biotinylated |
|
|
|
EpiDyne??Chromatin Remodeling? Assay Substrates | 16-4201 | EpiDyne FRET Nucleosome Remodeling? Assay Substrate |
16-4101 | EpiDyne Nucleosome Remodeling Assay? Substrate ST601-GATC1 |
16-4112 | EpiDyne Nucleosome Remodeling Assay? Substrate ST601-GATC1,2, biotinylated |
16-4113 | EpiDyne Nucleosome Remodeling Assay? Substrate ST601-GATC1,2,3, biotinylated |
16-4114 | EpiDyne Nucleosome Remodeling Assay? Substrate ST601-GATC1, 50-N-66, biotinylated |
16-4115 | EpiDyne Nucleosome Remodeling Assay? Substrate ST601-GATC1,2, 50-N-66, biotinylated |
16-4116 | EpiDyne Nucleosome Remodeling Assay? Substrate ST601-GATC1,2,3, 50-N-66, biotinylated |
|
|
|
SNAP Spike-in Controls | 19-1002 | SNAP-CUTANA? K-MetStat Panel |
19-1001 | SNAP-ChIP K-MetStat Panel |
19-2001 | SNAP-ChIP OncoStat Panel |
19-3001 | SNAP-ChIP K-AcylStat Panel |
|
|
|
dCypher? Nucleosome Panels | 16-9001 | dCypher? Nucleosome Full Panel |
?
想了解更多關(guān)于EpiCypher重組核小體技術(shù)和產(chǎn)品的信息嗎�����?請(qǐng)聯(lián)系欣博盛����!
?
EpiCypher的注冊(cè)商標(biāo)和知識(shí)產(chǎn)權(quán)可見(jiàn)鏈接:https://www.epicypher.com/intellectual-property/。
本文中的所有其他商標(biāo)和商品均為其各自公司所有���。
本文翻譯自鏈接:https://www.epicypher.com/resources/blog/finding-the-best-substrate-for-studying-histone-modifications/�����,如與原文有出入的地方����,請(qǐng)以英文原文為準(zhǔn)。
未經(jīng)EpiCypher公司事先書(shū)面同意����,本文件不得部分或全部復(fù)制。
?
關(guān)于EpiCypher公司:
EpiCypher是一家成立于2012年的表觀遺傳學(xué)公司����。從專(zhuān)有組蛋白肽陣列平臺(tái)EpiGold?開(kāi)始,EpiCypher開(kāi)發(fā)了一系列同類(lèi)產(chǎn)品���。同時(shí)�,EpiCypher是重組核小體制造和開(kāi)發(fā)的全球領(lǐng)導(dǎo)者���。利用其獨(dú)有技術(shù)���,不斷增加產(chǎn)品庫(kù)中高純度修飾重組核小體(dNucs?)產(chǎn)品。dNuc?多樣性的產(chǎn)品為破譯組蛋白編碼和加速藥物開(kāi)發(fā)提供了強(qiáng)大的工具�����。
EpiCypher還將dNuc?技術(shù)廣泛的應(yīng)用于多種分析測(cè)定產(chǎn)品中��,包括:SNAP-ChIP??Spike-in Controls(用于抗體分析和ChIP定量), EpiDyne??底物(用于染色質(zhì)重塑和抑制劑篩選及開(kāi)發(fā))�,dCyher?測(cè)定(用于探究表觀遺傳蛋白質(zhì)-組蛋白PTM結(jié)合相互作用)��。最近����,EpiCypher還推出了針對(duì)ChIC、CUT&RUN和CUT&Tag的高靈敏度表觀基因組圖譜CUTANA?分析����。
?
如需了解更多詳細(xì)信息或相關(guān)產(chǎn)品,請(qǐng)聯(lián)系EpiCypher中國(guó)授權(quán)代理商-欣博盛生物?
全國(guó)服務(wù)熱線: 4006-800-892? ? ? ?郵箱: market@neobioscience.com? ?
深圳: 0755-26755892? ? ? ?北京: 010-88594029? ? ? ? ?
上海: 021-34613729? ? ? ? ?廣州: 020-87615159? ? ? ? ??
代理品牌網(wǎng)站: m.smblzp.com? ?
自主品牌網(wǎng)站: www.neobioscience.net