SNAP-CUTANA? DYKDDDDK Tag Panel是 CUT&RUN 的加標(biāo)對(duì)照�����,提供了一種用于驗(yàn)證抗 FLAG?* 抗體的分析內(nèi)對(duì)照�,并證實(shí)了涉及FLAG表位標(biāo)記的染色質(zhì)蛋白的CUT&RUN反應(yīng)的成功。這種重要的陽(yáng)性對(duì)照可指導(dǎo)故障排除��,以區(qū)分 FLAG 表位標(biāo)記問(wèn)題(包括轉(zhuǎn)基因表達(dá)�、標(biāo)記蛋白的染色質(zhì)結(jié)合、標(biāo)簽的溶劑可及性等)與 CUT&RUN 工作流程中的技術(shù)故障�����。該panel由兩個(gè)包含未修飾組蛋白H3或3xDYKDDDDK-H3融合的核小體組成���,每個(gè)核小體都包裹有兩個(gè)獨(dú)特的條形碼DNA模板(A和B�,用于內(nèi)部技術(shù)復(fù)制)��。核小體單獨(dú)與順磁珠偶聯(lián)并匯集到單個(gè)panel中����,方便一步加標(biāo)到CUT&RUN反應(yīng)。在添加抗 DYKDDDDK 或 IgG 陰性對(duì)照抗體之前��,將 panel 與 ConA 固定的細(xì)胞一起添加(參見(jiàn)應(yīng)用說(shuō)明和表 1)。pAG-MNase釋放基因組染色質(zhì)和條形碼核小體取決于所使用抗體的特異性��。測(cè)序后��,回收的 DYKDDDDK 與未修飾核小體的相對(duì)讀取計(jì)數(shù)提供了靶向與脫靶回收率的定量指標(biāo)(圖 2)��,從而衡量實(shí)驗(yàn)成功率并指導(dǎo)故障排除工作��。有關(guān)工作流集成�、預(yù)期結(jié)果、數(shù)據(jù)分析和故障排除的詳細(xì)信息��,請(qǐng)參閱最新的CUTANA? CUT&RUN方法(https://www.epicypher.com/resources/protocols/cutana-cut-and-run-kit-manual)和SNAP-CUTANA? Spike-in(https://www.epicypher.com/resources/protocols/)用戶指南。
*FLAG? is a registered trademark of Merck KGaA, Darmstadt, Germany and ANTI-FLAG is a trademark of Sigma-Aldrich Co. LLC.
保存溫度
自收到之日起���,-20℃下可穩(wěn)定儲(chǔ)存6個(gè)月。較低的溫度會(huì)導(dǎo)致凍結(jié)����,并會(huì)永久損壞磁珠。
驗(yàn)證數(shù)據(jù)
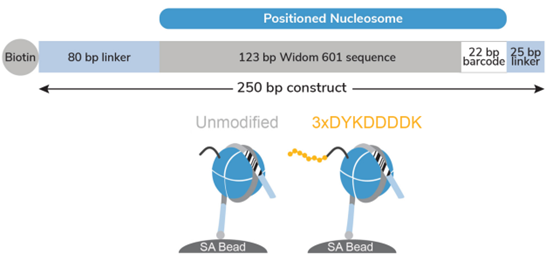
Figure 1: Schematic of SNAP-CUTANA? DYKDDDDK Tag Panel |
The DYKDDDDK Tag Panel contains two nucleosomes - one has an H3 tail fusion to a 3xDYKDDDDK Tag epitope and one is an unmodified control. Both octamers are wrapped with two uniquely barcoded DNA templates (A and B). Each 250 bp DNA template contains a 123 bp 601 nucleosome positioning sequence (gray) [1], a unique 22 bp DNA-barcode (white; 4 barcodes total), and a 5’ biotin-TEG. The 5’ and 3’ linkers (blue) are compatible with cleavage by pAG-MNase (EpiCypher?14-1048,?15-1016) during CUT&RUN. The nucleosomes are individually pre-conjugated to paramagnetic beads and pooled for convenient use.
|
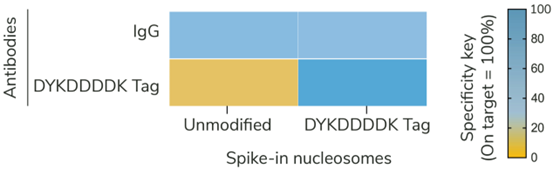
Figure 2: SNAP-CUTANA? DYKDDDDK Tag Panel provides an in-assay control for CUT&RUN? reactions targeting FLAG-tagged proteins |
CUT&RUN was performed as described in?Figure 5. CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the SNAP-CUTANA? DYKDDDDK Tag Panel. Data are expressed as a percent relative to on-target recovery (DYKDDDDK Tag set to 100%) or total counts (IgG). IgG antibody results demonstrate equal loading of unmodified and epitope nucleosomes in the panel. DYKDDDDK Tag antibody results show selective enrichment of the DYKDDDDK Tag spike-in nucleosomes, validating all CUT&RUN steps, including DYKDDDDK antibody binding, pAG-MNase cleavage, and wash conditions.
|
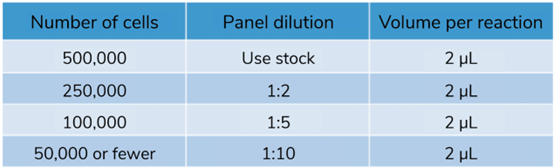
Table 1: Recommended SNAP-CUTANA? DYKDDDDK Tag Panel Spike-in dilution for CUT&RUN reactions of varying starting cell number. |
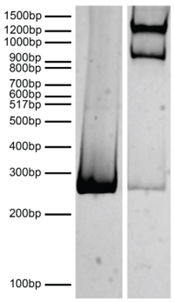
Figure 3: DNA gel data |
Nucleosomes in the SNAP-CUTANA? DYKDDDDK Tag Panel were resolved via native PAGE and stained with ethidium bromide to confirm intact nucleosome assembly.?Lane 1:?Free 250 bp DNA used in nucleosome assembly (100 ng).?Lane 2:?Intact nucleosomes (400 ng).
|
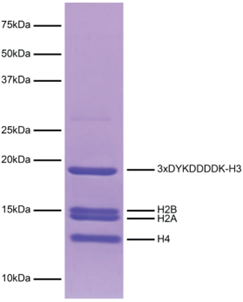
Figure 4: Protein gel data |
Coomassie stained SDS-PAGE gel of the nucleosome containing a 3xDYKDDDDK-H3 fusion (1 μg) in the SNAP-CUTANA DYKDDDDK Tag Panel demonstrates the purity of histones in the preparation. Sizes of molecular weight markers and positions of the core histones (H2A, H2B, 3xDYKDDDDK-H3, and H4) are indicated.
|
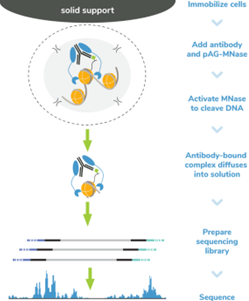
Figure 5: CUT&RUN methods |
CUT&RUN was performed on 500k MDA-MB-231 native cells stably expressing 3xFLAG-tagged GATA3 [1]** using the CUTANA? ChIC/CUT&RUN Kit v3 (EpiCypher?14-1048). SNAP-CUTANA? DYKDDDDK Tag Panel was added just prior to the addition of either DYKDDDDK Tag (0.05 μg; EpiCypher?13-2031) or IgG negative control (0.5 μg; EpiCypher?13-0042) antibodies. Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA? CUT&RUN Library Prep Kit (EpiCypher?14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
**Thanks to Dr. Takaku (UND) for 3xFLAG-GATA3-3xHA MDA-MB-231 cells. |
訂購(gòu)詳情
貨號(hào) | 產(chǎn)品名稱 | 規(guī)格 |
19-5001 | SNAP-CUTANA? DYKDDDDK Tag Panel | 50 Reactions |
參考文獻(xiàn)
[1] Lowary & Widom J. Mol. Biol. (1998). PMID: 9514715
[2] Takaku et al. Genome Biol. (2016). PMID: 26922637
?

如需了解更多詳細(xì)信息或相關(guān)產(chǎn)品����,
請(qǐng)聯(lián)系EpiCypher中國(guó)授權(quán)代理商-欣博盛生物?
全國(guó)服務(wù)熱線: 4006-800-892 ? ? ??郵箱: market@neobioscience.com?
深圳: 0755-26755892 ? ? ? ??北京: 010-88594029 ???????????
廣州:020-87615159????????? ?上海: 021-34613729
代理品牌網(wǎng)站: m.smblzp.com?
自主品牌網(wǎng)站: www.neobioscience.net